| J Korean Neurol Assoc > Volume 42(2); 2024 > Article |
|
Abstract
Background
Charcot-Marie-Tooth disease type 1A (CMT1A) is caused by duplication of the 17p12 region including PMP22 gene. In CMT1A patients, anticipation showing increased severity by generations has been reported in the CMT1A patients. It has also been reported that severity increases in the non-de novo cases than in the de novo cases. This study was performed to examine epigenetic differences between CMT1A cases and controls as well as between de novo cases and non-de novo cases.
Methods
This study examined 40 Korean CMT1A patients and 11 controls. Methylation level was determined using the SureSelect XT Methyl-Seq reagent kit and bisulfite sequence mapping program.
샤르코-마리-투스병(Charcot-Marie-Tooth disease, CMT)은 난치성 말초신경병으로 임상 증상과 유전적 발병 원인이 이질적인 희귀 유전병에 속한다. 지금까지 약 130개 이상의 유전자의 변이가 CMT를 유발하는 것으로 보고되었다[1,2]. 그중에서도 PMP22 유전자를 포함하는 17번 염색체 17p12 부위의 1.4 Mb 길이에 대한 불균등 교차(unequal crossover)로 인한 중복(duplication)과 결손(deletion)은 CMT 1A형(CMT1A)과 압박마비취약유전신경병(hereditary neuropathy with liability to pressure palsies)을 각각 유발한다[3,4]. CMT1A는 CMT 환자들 중 약 50-70% 정도로 매우 높은 빈도로 발생한다[5,6].
몇몇 CMT1A 가계에서 세대가 거듭될수록 환자들의 빠른 발병 연령과 중증화 경향의 대물림 악화(anticipation)가 보고되었다[7]. Steiner 등[7]은 de novo 돌연변이로 CMT1A가 발병한 부모 세대에서 평균 발병 연령은 41.2세인 반면 돌연변이를 물려받은 다음 세대 자식들은 12.6세로 유의하게 더 빨랐으며 중증도 또한 자식 세대에서 더 심하다는 것을 보고하였다. 다른 연구에서도 비슷한 결과를 보고하였는데 Lee 등[8]은 CMT 신경병 점수(CMT neuropathy score)로 측정된 중증도가 de novo CMT1A 환자에 비해 non-de novo 환자가 더 심했으며 출생 시 de novo군의 아버지의 연령이 non-de novo군에 비해 약간 높다고 보고하였다. 아울러 미발표 결과 이긴 하나 Beck 우울증 검사(Beck depression inventory)에서도 de novo CMT1A 환자들이 non-de novo군에 비해 낮은 값을 보여 덜 우울하다는 조사 결과도 있었다. Lee 등[8]은 de novo 환자군이 non-de novo 환자군에 비해 증상이 약하거나 발병 연령이 늦어지는 기전을 규명하지는 못했지만 후성유전학 요인이 작용하였을 가능성을 제시하였다.
후성유전학적으로 DNA의 CpG 서열의 메틸화는 유전자 발현과 세포 분화의 조절에 중요한 역할을 한다[9]. DNA 메틸화는 불안장애를 비롯한 많은 정신장애에 관여하는 것으로 알려져 있다[10]. 아동기의 스트레스는 메틸화의 차이를 유발하기도 한다. 어린 시절의 만성적이고 심각한 스트레스 노출 정도를 평가한 연구에서 550개의 스트레스 관련 차등 메틸화 CpG 부위(differentially methylated CpG site, DMC)를 포함하는 122개 유전자를 발견하였으며 차등 유전자 발현을 보이는 12개 유전자와 DMC의 연관 유전자에서 동시에 발견되어 메틸화가 유전자 발현에 영향을 주었을 것으로 예상하였다[11]. 어린 원숭이에서 불안 및 우울증의 위험인자(risk factor)와 연관된 유전자들의 차등 메틸화가 불안 행동과 관련이 있는 것으로 보고되기도 하였다[12]. 또한 동물 모델의 실험에서는 스트레스 노출이 다음 세대까지 후성유전학 변화를 초래함으로써 외상(trauma)에 대한 자손의 증상을 심화하거나 불안이나 우울증이 다음 세대로 전달될 수 있다는 잠재적 가능성이 제시되었다[11,13,14].
본 연구에서는 동일한 유전적 원인으로 발병한 CMT1A 환자들을 대상으로 대조군에 대한 후성유전학적 차이를 먼저 조사하였으며 de novo 환자군과 non-de novo 환자군에서 발병 연령 및 중증도의 차이와 후성유전학 차이 간의 연관성을 분석하였다.
본 연구는 PMP22를 포함하는 17p12 부위의 중복이 확인된 40명의 한국인 CMT1A 환자(증례군)와 11명의 정상 대조군을 대상으로 수행하였다(Table 1). 증례군과 대조군의 메틸화 비교를 위해서는 30-40대 남성의 CMT1A 환자 20명과 대조군 11명을 대상으로 분석을 수행하였다. CMT1A 가계를 대상으로 세대 간 환자들의 메틸화 비교를 위해서는 부모가 모두 비환자인 de novo 환자 10명과 부모 중 한 명이 환자인 non-de novo 환자 10명으로 군을 나누어 분석을 수행하였다. 모든 연구 참가자들은 삼성서울병원에서 모집하였고 서면 동의서를 받았으며 삼성서울병원(2014-08-057-002)과 공주대학교(KNU_IRB_2018-06)의 기관생명윤리위원회의 승인을 받았다.
환자의 중증도는 기능장애 척도(functional disability scale, FDS)와 CMT 신경병 점수 2판(CMT neuropathy score version 2, CMTNSv2)으로 측정하였다. 발병 연령은 환자에게 원위부근육 약화, 발 기형, 감각 변화 등의 증상이 처음 나타난 나이를 추적하여 결정하였다. 정중운동신경 전도 속도(median motor nerve conduction velocity, MNCV)는 팔꿈치와 손목에 자극을 준 후 측정하였다.
말초 혈액으로부터 QIAamp DNA mini kit (Qiagen, Hilden, Germany)를 사용하여 유전체(genomic) DNA를 추출하였다. CMT1A의 발병 원인인 17p12의 복제수 변이는 1.4 Mb 영역에 분포하는 여섯 개의 미세부수체(microsatellite)의 유전자형 결정과 quantitative polymerase chain reaction을 통한 PMP22 유전자의 유전체 내 정량으로 결정하였다. De novo 돌연변이 환자 가계의 친자 여부는 PowerPlex Fusion System (Promega, Madison, WI, USA)을 사용하여 short tandem repeats (STRs)의 유전자형의 비교를 통해 확인하였다.
메틸화 수준은 SureSelect XT Methyl-Seq reagent kit (Illumina, San Diego, CA, USA)로 라이브러리를 제작하고 Illumina platform sequencer (Illumina)를 사용하여 결정하였다. Methylseq 데이터 분석은 hg19 참조서열을 기준으로 Bisulfite Sequence Mapping Program (http://code.google.com/p/bsmap/)을 사용하여 모든 유전자의 표적 농축 부위(target-enriched regions)에 대한 메틸화 수준을 측정하였다. 메틸화 수준의 정량화를 위해서는 CpG 서열에 대한 C 수/C+T 수로 계산하고 중앙값 스케일링 표준화를 사용하여 데이터를 정규화하였으며 실험 오류를 배제하기 위해 판독(read) 수가 500개 이상이거나 10개 이하인 CpG 부위는 제외한 후 통계값을 산출하였다(10<read<500). 모든 유전자의 프로모터(promoter)는 전사 개시 지점으로부터 2 kb 상류 부위로 정의하였다.
통계 분석은 Python (Python Software Foundation, Fredericksburg, VA, USA)의 통계 분석 라이브러리인 SciPy를 이용하여 수행하였다[15]. 각 CpG 부위의 군별 메틸화 정도에 대한 분석은 독립 t-test와 계통 집락화(hierarchical clustering) 방법을 사용하였고 히트맵(heatmap)은 유클리드 거리(Euclidean distance)와 완전 연관법(complete linkage)을 적용하여 작성하였다. 군들의 임상정보 분석 및 그래프는 GraphPad Prism version 8.00 (GraphPad Software, Boston, MA, USA)과 OriginPro 2018 (OriginLab Corporation, Northampton, MA, USA)을 사용하여 작성하였다. DMC와 관련된 유전자의 기능에 대한 분석은 상염색체에 위치한 DMC와 연관된 유전자를 대상으로 수행하였다. 정신적 스트레스 또는 신경에 영향을 주는 유전자에서 메틸화 차이를 유발한다고 보고된 CpG 부위에 대한 정보는 epigenome-wide association study (EWAS) Atlas (https://ngdc.cncb.ac.cn/ewas/atlas)를 참조하였다. 비교군 간 메틸화 차이가 p<0.01인 CpG 자리와 연관된 유전자 세트에 대해 Enrichr (https://maayanlab.cloud/Enrichr/)에서 gene ontology (GO, http://geneontology.org/)를 기반으로 3가지 GO 분야(생물학 과정, 분자기능, 세포 구성)에 대해 GO term enrichment를 수행하여 해당 DMC가 위치한 유전자가 어떤 분야 또는 경로에 관여하는지를 조사하였다. 확률(p)은 hypergeometric test 또는 Fisher’s exact test로구했으며 유전자의 확률에 대한 이항분포 및 독립성을 가정하였다. GO 분야에 주석이 달린 유전자 중에서 DMC를 포함하는 유전자들의 비율이 높아 p<0.05로 유의하게 예측된 항목 중 상위 10개 분야로 그래프를 작성하였다.
본 연구는 11명의 대조군과 20명의 CMT1A 증례군의 비교 및 각 10명으로 구성된 de novo CMT1A군과 non-de novo CMT1A군의 군 간 비교를 각각 독립적으로 수행하였다(Table 1). 모두 남성으로만 구성된 대조군과 증례군의 첫 번째 비교에서 검사 당시 평균 연령은 각각 42.2±3.4세와 42.6±3.0세로 유의한 차이가 없었다. 6명의 남성과 4명의 여성으로 동일한 남녀 비율로 구성된 de novo CMT1A군과 nonde novo CMT1A군의 두 번째 비교에서 평균 연령은 26.7± 14.9세와 24.5±9.1세로 비교군 간 연령은 유의한 차이가 없었으며(p=0.694) 평균 발병 연령은 18.7±13.8세와 9.6±3.0세로 de novo군이 9.1세 정도 늦은 발병을 보였고(p=0.057) 평균 질병 기간은 8.0±8.9세와 14.9±9.4세로 de novo군에서 유의하게 짧았다(p=0.027). 평균 CMTNSv2는 de novo군과 non-de novo군에서 각각 7.3±2.9점과 14.0±4.4점으로 non-de novo군에서 유의하게 증상이 심했으며(p=0.001) 평균 FDS는 각각 1.2±0.4점과 1.8±0.9점으로 non-de novo군에서 약간 높았지만 유의한 차이는 없었다(p=0.141). 평균 MNCV는 각각 19.5±5.1 m/s와 18.1±3.2 m/s로 두 군 간에 유의한 차이를 보이지 않았다(p=0.141). 본 연구에서 관찰된 de novo군과 non-de novo군 사이의 임상 차이는 Lee 등[8]의 연구와 일치하였다.
각 그룹 간 비교에서 전체 6,366,410 CpG에 대해 메틸화 수준을 분석하였으며 비교군 내 정규화를 통해 바이어스(bias)를 제외한 2,301,420 CpG에 대해서 통계 분석을 수행하였다. 성별에 의한 차이를 배제하고자 성염색체를 제외한 상염색체 CpG만을 비교 대상으로 하였다. 대조군과 증례군을 대상으로 한 비교에서 p<0.05를 충족하는 114,384개 DMC와 p<0.05와 |Δmean|≥0.2를 동시에 충족하는 414개 DMC를 선별하였다(Fig. 1-A, C). 이들 중 211개 DMC가 대조군에 비해 증례군에서 고메틸화되었고 203개 DMC는 증례군에서 저메틸화되었다.
De novo군과 non-de novo군 간의 비교에서도 정규화 과정을 거쳐 3,635,687 CpG의 메틸화 수준에 대해 통계 분석을 수행하여 p<0.05를 충족하는 111,570개 DMC와 p<0.05와 |Δmean|≥0.2를 동시에 충족하는 1,055개 DMC를 선별하였다(Fig. 1-B, D). 선별된 DMC 중 590개는 non-de novo군에서 고메틸화되었으며 465개는 저메틸화되었다.
상기의 두 비교에서 p<0.05와 |Δmean|≥0.2를 동시에 만족하는 414개 CpG와 1,055개 CpG를 대상으로 heatmap을 제작하였을 때 각 군별로 바르게 집락화되었음을 볼 수 있었다(Fig. 2).
비교군 간 메틸화의 차이(p<0.05)를 보이면서 프로모터나 유전자 내부에 존재하는 DMC들의 특성에 대해 조사하였다. 증례군과 대조군 간의 비교에서 p<1×10-5인 DMC는 20개가 분리되었고(Table 2) 분리된 20개의 DMC 중 증례군에서의 고메틸화(positive)는 7개, 저메틸화(negative)는 13개로 나타났다. 유전자의 프로모터 부위에 위치한 DMC는 6개였으며 1개는 5’ untranslated region (5’ UTR) 지역, 4개는 엑손, 6개는 인트론, 3개는 유전자 간(intergenic) 지역, 1개는 ncRNA의 인트론으로 23개의 유전자를 포함하는 위치에 있었다. De novo군과 non-de novo군의 비교에서 p<1×10-5인 DMC는 9개가 분리되었는데 non-de novo 환자군에서 4개는 고메틸화, 5개는 저메틸화 양상을 보였다. 유전자의 프로모터 부위에 위치한 DMC는 3개였으며 1개는 5’ UTR 지역, 1개는 엑손, 2개는 인트론, 2개는 유전자 간 부위로 11개의 유전자를 포함하는 위치에 있었다.
CMT를 발병하는 돌연변이로 알려진 DYNC1H1 유전자의 프로모터 부위와 IGHMBP2 유전자 인트론의 DMC가 대조군보다 증례군에서 고메틸화되었다. LHPP, KLHDC1, HTR5A는 정신질환 관련 유전자[16-18]로 해당 유전자 주변에 위치한 DMC가 두 비교군에서 여러 개 분리되었다. 대조군과 증례군의 비교에서 LHPP와 FAM53B 유전자 사이에 위치한 DMC와 KLHDC1의 프로모터에 위치한 DMC가 연관이 있었다. LHPP와 연관된 DMC는 증례군에서 대조군보다 고메틸화되었으며 KLHDC1와 연관된 DMC는 저메틸화되었다. De novo군과 non-de novo군의 비교에서 CMT를 일으키는 유전자 주변의 DMC에서는 메틸화 수준의 차이는 없었지만 HTR5A의 프로모터에 위치한 DMC에서 non-de novo군이 de novo군에 비해 고메틸화되었다.
모든 DMC에 대해 EWAS 데이터에서 이전에 보고된 동일한 위치 중 정신적 스트레스 또는 운동능력과 신경전달물질에 영향을 준다고 보고된 특성의 CpG 자리에 대해 조사하였다. 증례군과 대조군 비교에서 6개, de novo군과 non-de novo군의 비교에서 8개의 CpG가 연관이 있는 것으로 나타났다(Tables 3, 4). 두 비교에서 공통적으로 선별된 CpG는 없었으나 걸프전병(Gulf War illness)과 아동 학대와 연관된 CpG 자리가 공통적으로 분리되었다[19,20].
증례군과 대조군 비교에서 파킨슨병[21], 다발경화증[22], 걸프전병[19], 펠란-맥더미드증후군(Phelan-McDermid syndrome) [23], 조기사회박탈감[24], 아동 학대[20]에 관한 연구에서의 CpG 위치와 증례군과 대조군의 DMC의 위치가 일치하였다(Table 3). 그중 3개 CpG 자리(cg11963436, cg09015566, cg03259243)는 대조군에 비해 증례군에서 고메틸화되었고[19-21] 1개 CpG 자리(cg21006834)는 저메틸화되어 이전에 보고된 양상과 일치하였다[23].
De novo군과 non-de novo군의 비교에서는 조현병(schizophrenia) [25], 아동기 스트레스[11], 아동 학대[20], 걸프전병[19], 운동 전후[26], 강직척추염[27]에 관한 연구에서 CpG의 위치가 일치하였다(Table 4). 그중 5개 CpG (cg18303477, cg03144560, cg23144563, cg17295225, cg02487331)는 de novo군에 비해 non-de novo군에서 고메틸화되었고[11,19,20,28] 1개 CpG 자리(cg05828624)는 저메틸화되어 이전에 보고된 현상과 일치하였다[26].
DMC가 내외에 위치한 유전자를 세트로 사용하여 GO enrichment analysis를 수행하였다. GO에서 제공하는 3가지 항목인 생물학 과정, 분자기능, 세포 구성 요소에 대해 p-value를 기준으로 상위 10개 GO 분야를 그래프로 나타내었다(Fig. 3). 증례군과 대조군 간 비교에서 생물학 과정의 분야 중 신경세포 생성(p<1×10-8)과 신경세포 분화(p<1×10-7), 신경계 발달(p<1×10-6) 범주가 상위에서 가장 유의하였다(Fig. 3-A). De novo군과 non-de novo군에서는 신경세포 분화(p<1×10-5)와 배아기관 형성(p<1×10-5), 신경세포 생성(p<1×10-4) 범주에서 유의하였다(Fig. 3-B). 두 군 간 비교에서 모두 신경세포 분화 및 생성, 신경계 발달과 관련된 범주가 공통적으로 분류되었다.
분자기능을 담당하는 유전자 분야는 두 군 간의 비교에서 모두 상위 3개 범주가 공통적으로 나타났다. 이는 모두 DNA 결합 관련 유전자로 DNA 서열-특이 결합, 서열-특이 이중가닥 DNA 결합, 이중가닥 DNA 결합에 관여하는 유전자가 가장 유의하게 많은 부분을 차지했다(p<1×10-5).
세포 구성에 따라 분류한 항목은 모두 1×10-3<p<1×10-1로 다른 분류군에 비해 유의성이 낮았다. 증례군과 대조군 간의 비교에서 가장 유의하게 많은 유전자를 차지한 항목은 polycomb repressive complex 1 (PRC1)이었고 칼슘이온통로 복합체, 신경세포 돌출(neuron projection) 등 신경과 관련된 유전자 비율이 높은 것으로 나타났다. De novo군과 non-de novo군의 비교에서는 이온성 글루탐산염(glutamate) 수용체 복합체가 가장 높은 비율로 유의성을 나타냈고 세포연접과 관련된 유전자가 그 다음을 차지하였다. 두 비교군에서 공통적으로 유의한 유전자 범주는 수상 돌기와 근육세포의 외막 구성에 관여하는 유전자군으로 나타났다.
본 연구는 한국의 CMT1A 환자를 대상으로 증례군과 대조군 그리고 de novo군과 non-de novo군이 동일한 PMP22 유전자의 중복으로 발병하였으나 중증도의 차이를 나타내는 원인을 규명하기 위해 DNA 메틸화 양상을 분석하였다. 증례군과 대조군에서 DMC가 위치한 주변 유전자 중 DYNC1H1과 IGHMBP2는 CMT를 일으킨다고 알려져 있으며[28,29] ADCY5 돌연변이는 근긴장운동이상을 일으키고[30] KLHDC1는 전유전체 연관 분석(genome wide association study)에서 여러 변이들의 단상형(haplotype) 조합이 조울증을 유발하는 것으로 보고되었다[17]. De novo군과 non-de novo군의 DMC가 위치한 HTR5A의 돌연변이는 주요 우울장애(major depressive disorder, MDD)를 유발하는 것으로 알려져 있다[16]. LHPP와 KLHDC1은 두 비교군 모두 공통적으로 유의한 DMC 연관 유전자로 LHPP는 생쥐에서 스트레스우울증의 조절인자로 작용하며[31] 사람에서는 LHPP 유전자 변이에 따라 MDD에 영향을 미치는 것으로 알려져 있다[18]. 본 연구 결과에서 증례군과 대조군, de novo군과 non-de novo군의 비교를 보면 모두 동일한 CpG 부위에서 유의성을 보이지는 않았으나 신경 및 관련 질병에 연관된 유전자에서 메틸화 수준의 차이를 보이는 것은 신경이상 혹은 지속 우울증으로 인해 후성유 전학적 변화를 초래했을 가능성이 있다.
2023년 기준 EWAS Atlas에 활용된 1,002편의 EWAS 논문을 기반으로 728개의 특성들에 대한 정보를 바탕으로 신경계 혹은 외상이나 스트레스와 연관성을 비교 분석하였다. 정신적 스트레스 연관 특성은 걸프전병[19]과 아동 학대[20]가 두 비교군에서 공통적으로 나타났다. 걸프전병은 1991년 걸프전에 참여한 퇴역 군인에게서 공통적으로 발견되는 신경질환으로 독성물질에 대한 노출 및 정신적 스트레스에 의해 신경계기능장애를 나타냈다[32]. 특히 아동 학대를 겪는 어린이군과 그렇지 않은 어린이군 간의 차이를 분석한 연구에서 학대를 받은 군에서 cg03259243, cg03144560, cg23144563 CpG들이 고메틸화되었는데[20] 본 연구에서도 증례군 및 non-de novo군에서 모두 동일한 메틸화 경향을 보였다. 공통적으로 나타나는 특성을 고려했을 때 유전병 발병에 의한 정신적 스트레스가 환자들에게 후성유전학적 변화를 유발했을 가능성이 제기된다. 반면 cg07443748, cg25040670, cg07875360, cg13904130의 CpG 자리에서는 이전 보고들과는 다른 양상의 메틸화 차이를 보였는데 DNA 메틸화의 변화가 세포의 단계 및 조직 특이적 영향을 받기 때문에 나타난 결과일 수 있다. 실제 cg07443748는 CD19+ B-cell [22], cg18303477는 타액에서 분석되었으며[11] 사용된 검체의 종류 이외에도 인종이나 자라온 환경 등 기타 다른 요소에 의한 차이일 가능성도 있다.
본 연구의 두 비교군에서의 p<0.01인 DMC를 포함하는 주변 유전자들에 대한 기능 분석에서 모두 신경세포와 시냅스의 발달 및 분화, 이온통로 등 신경계와 연관된 여러 분야에서 높은 유의성을 확인하였다. 또한 신경세포의 생성과 분화에 관여하는 범주가 증례군과 대조군과의 비교가 de novo군과 non-de novo군 비교에 비해 p-value가 더 유의하고 유전자 수가 많았다. 이것은 신경세포의 주요 기능 유전자의 DNA 메틸화가 질병과 연관된 유전자 발현에 영향을 줄 수 있다는 것을 의미하며 신경질환에서 신경세포 연관 유전자의 DNA 메틸화는 질병의 중증도에 영향을 줄 수 있다.
신경전달물질과 관련된 단백질을 암호화하는 유전자가 많이 확인되었는데 증례군과 대조군과의 비교에서 글루탐산염(glutamate) 수용체 신호전달과정, 타이로신인산화효소(tyrosine kinase) 활성 및 수용체에 관여하는 유전자가 해당된다. De novo군과 non-de novo군 간의 비교에서는 글루탐산염 및 아드레날린 수용체 결합 등 시냅스전달물질 관련 유전자가 높은 비율을 차지하였다. 타이로신은 아드레날린과 같은 흥분성 전달물질의 대사 전구체로 작용하는 도파민의 초기 생합성 단계에 필요하며 뇌기능에 중요한 역할을 한다[33]. 그리고 글루탐산염은 많은 신경조직에 존재하는 주요 흥분성 신경전달물질로 우울증에 대한 치료제로 활용되기도 한다[34]. 글루탐산염은 중추신경계의 주요 신경전달 억제물질인 감마아미노뷰티르산(γ-aminobutyric acid, GABA) 합성에 사용되며 GABA는 신경계 전반에 걸쳐 흥분성을 조절하는 역할을 하여 근긴장 조절에 직접 역할을 한다[35]. 이러한 요인들은 환자에서 항상성을 유지하기 위한 전사 조절 및 신경전달물질에 관여하는 유전자의 후성유전학적 변화는 세대에 따라 증상이 더욱 심해지거나[8] 더 강한 정신적 스트레스로 인해 de novo군과 non-de novo군 간의 차이를 초래하였을 가능성을 제시한다. 세포 구성에 관여하는 유전자의 분류에서는 두 비교군에서 공통적으로 수상돌기, 골격근세포의 세포막을 구성하는 다수 유전자의 DNA 메틸화 차이가 관찰되었다. 이는 환자뿐만 아니라 상대적으로 중증인 non-de novo 그룹에서 신경 관련 유전자의 발현에 영향을 주었을 가능성을 제기한다. 지금까지 신경정신질환과 관련된 후성유전학적 연구들은 우울증과 같이 다인자적 요인으로 발병하는 질병이나 충격적인 사건을 겪은 후 외상을 가진 사람들을 대상으로 한 연구가 대부분이었다. 본 연구는 1차적으로 유전적 발병 원인이 명확히 밝혀진 멘델 유전의 CMT1A 말초신경병을 대상으로 했다는 점에서 기존 연구들과 차이가 있다. 본 연구를 통해 증례군과 대조군 그리고 de novo군과 non-de novo군 사이의 비교에서 신경세포 및 신경전달물질과 관련된 유전자들에서 메틸화 차이가 다수 발견되었다는 것은 CMT1A에 대한 중증도를 조절하는 후성유전학적 요인이 있음을 암시한다. 이러한 결과는 환경 요인이나 스트레스에 따른 후성유전학적 변화의 차이는 CMT 환자들의 임상 이질성 혹은 세대를 거듭할수록 증상이 심해지는 유전 요인에 대한 연구의 시발점이 될 수 있음을 시사한다. 아픈 부모를 보며 자라온 자녀의 생활 환경과 정신적 스트레스 및 다양한 복합 요인의 상호작용은 신경전달물질이나 이온통로 연관 유전자에서 메틸화 차이를 유발했을 수 있다. 본 연구는 희귀 유전병을 대상으로 하였기 때문에 제한된 수의 환자를 대상으로 수행되었다는 한계가 있지만 CMT 환자들을 대상으로 발병 연령 및 중증도 이질성의 원인을 찾기 위한 첫 후 성유전학 연구로써 가치가 있다고 판단된다.
Acknowledgements
This research was supported by the National Research Foundation of Korea grant, funded by the Ministry of Education, Science and Technology (NRF-2019R 1A2C1087547, NRF-2020M3H4A1A03084600, NRF-2021R1A4A2001389, and NRF-2022M3E5E9016662).
REFERENCES
1. Pipis M, Rossor AM, Laura M, Reilly MM. Next-generation sequencing in Charcot-Marie-Tooth disease: opportunities and challenges. Nat Rev Neurol 2019;15:644-656.


2. Rossor AM, Carr AS, Devine H, Chandrashekar H, Pelayo-Negro AL, Pareyson D, et al. Peripheral neuropathy in complex inherited diseases: an approach to diagnosis. J Neurol Neurosurg Psychiatry 2017;88:846-863.


3. Lupski JR, de Oca-Luna RM, Slaugenhaupt S, Pentao L, Guzzetta V, Trask BJ, et al. DNA duplication associated with Charcot-MarieTooth disease type 1A. Cell 1991;66:219-232.


4. Chance PF, Alderson MK, Leppig KA, Lensch MW, Matsunami N, Smith B, et al. DNA deletion associated with hereditary neuropathy with liability to pressure palsies. Cell 1993;72:143-151.


5. Rudnik‐Schöneborn S, Thiele S, Walter MC, Reinecke L, Sereda M, Schöneborn R, et al. Pregnancy outcome in Charcot-Marie-Tooth disease: results of the CMT-NET cohort study in Germany. Eur J Neurol 2020;27:1390-1396.



6. Gemelli C, Geroldi A, Massucco S, Trevisan L, Callegari I, Marinelli L, et al. Genetic workup for Charcot-Marie-Tooth neuropathy: a retrospective single-site experience covering 15 years. Life 2022;12:402.



7. Steiner I, Gotkine M, Steiner-Birmanns B, Biran I, Silverstein S, Abeliovich D, et al. Increased severity over generations of Charcot-Marie-Tooth disease type 1A. J Neurol 2008;255:813-819.


8. Lee AJ, Nam DE, Choi YJ, Noh SW, Nam SH, Lee HJ, et al. Paternal gender specificity and mild phenotypes in Charcot-Marie-Tooth type 1A patients with de novo 17p12 rearrangements. Mol Genet Genomic Med 2020;8:e1380.



9. Rando OJ, Verstrepen KJ. Timescales of genetic and epigenetic inheritance. Cell 2007;128:655-668.


11. Papale LA, Seltzer LJ, Madrid A, Pollak SD, Alisch RS. Differentially methylated genes in saliva are linked to childhood stress. Sci Rep 2018;8:10785.



12. Alisch RS, Chopra P, Fox AS, Chen K, White AT, Roseboom PH, et al. Differentially methylated plasticity genes in the amygdala of young primates are linked to anxious temperament, an at risk phenotype for anxiety and depressive disorders. J Neurosci 2014;34:15548-15556.



13. Stenz L, Schechter DS, Serpa SR, Paoloni-Giacobino A. Intergenerational transmission of DNA methylation signatures associated with early life stress. Curr Genomics 2018;19:665-675.



14. Yehuda R, Bierer LM. The relevance of epigenetics to PTSD: implications for the DSM-V. J Trauma Stress 2009;22:427-434.



15. Virtanen P, Gommers R, Oliphant TE, Haberland M, Reddy T, Cournapeau D, et al. SciPy 1.0: fundamental algorithms for scientific computing in Python. Nat Methods 2020;17:261-272.


16. Yosifova A, Mushiroda T, Stoianov D, Vazharova R, Dimova I, Karachanak S, et al. Case-control association study of 65 candidate genes revealed a possible association of a SNP of HTR5A to be a factor susceptible to bipolar disease in Bulgarian population. J Affect Disord 2009;117:87-97.


17. Feng T, Zhu X. Genome-wide searching of rare genetic variants in WTCCC data. Hum Genet 2010;128:269-280.



18. Cui L, Gong X, Tang Y, Kong L, Chang M, Geng H, et al. Relationship between the LHPP gene polymorphism and restingstate brain activity in major depressive disorder. Neural Plast 2016;2016:9162590.



19. Trivedi MS, Abreu MM, Sarria L, Rose N, Ahmed N, Beljanski V, et al. Alterations in DNA methylation status associated with gulf war illness. DNA Cell Biol 2019;38:561-571.


20. Yang BZ, Zhang H, Ge W, Weder N, Douglas-Palumberi H, Perepletchikova F, et al. Child abuse and epigenetic mechanisms of disease risk. Am J Prev Med 2013;44:101-107.



21. Moore K, McKnight AJ, Craig D, O’Neill F. Epigenome-wide association study for Parkinson’s disease. Neuromolecular Med 2014;16:845-855.


22. Maltby VE, Lea RA, Graves MC, Sanders KA, Benton MC, Tajouri L, et al. Genome-wide DNA methylation changes in CD19+ B cells from relapsing-remitting multiple sclerosis patients. Sci Rep 2018;8:17418.



23. Schenkel LC, Aref-Eshghi E, Rooney K, Kerkhof J, Levy MA, McConkey H, et al. DNA methylation epi-signature is associated with two molecularly and phenotypically distinct clinical subtypes of Phelan-McDermid syndrome. Clin Epigenetics 2021;13:2.



24. Naumova OY, Rychkov SY, Kornilov SA, Odintsova VV, Anikina VО, Solodunova MY, et al. Effects of early social deprivation on epigenetic statuses and adaptive behavior of young children: a study based on a cohort of institutionalized infants and toddlers. PLoS One 2019;14:e0214285.



25. Jaffe AE, Gao Y, Deep-Soboslay A, Tao R, Hyde TM, Weinberger DR, et al. Mapping DNA methylation across development, genotype and schizophrenia in the human frontal cortex. Nat Neurosci 2016;19:40-47.



26. Lindholm ME, Marabita F, Gomez-Cabrero D, Rundqvist H, Ekström TJ, Tegnér J, et al. An integrative analysis reveals coordinated reprogramming of the epigenome and the transcriptome in human skeletal muscle after training. Epigenetics 2014;9:1557-1569.



27. Coit P, Kaushik P, Caplan L, Kerr GS, Walsh JA, Dubreuil M, et al. Genome-wide DNA methylation analysis in ankylosing spondylitis identifies HLA-B* 27 dependent and independent DNA methylation changes in whole blood. J Autoimmun 2019;102:126-132.


28. Corti S, Locatelli F, Papadimitriou D, Donadoni C, Del Bo R, Crimi M, et al. Transplanted ALDHhiSSClo neural stem cells generate motor neurons and delay disease progression of nmd mice, an animal model of SMARD1. Hum Mol Genet 2006;15:167-187.


29. Braunstein KE, Eschbach J, Ròna-Vörös K, Soylu R, Mikrouli E, Larmet Y, et al. A point mutation in the dynein heavy chain gene leads to striatal atrophy and compromises neurite outgrowth of striatal neurons. Human Mol Genet 2010;19:4385-4398.



30. Chen DH, Méneret A, Friedman JR, Korvatska O, Gad A, Bonkowski ES, et al. ADCY5-related dyskinesia: broader spectrum and genotype-phenotype correlations. Neurology 2015;85:2026-2035.



31. Lin D, Li L, Chen WB, Chen J, Ren D, Zheng ZH, et al. LHPP, a risk factor for major depressive disorder, regulates stress-induced depression-like behaviors through its histidine phosphatase activity. Mol Psychiatry 2023;28:908-918.


32. White RF, Steele L, O’Callaghan JP, Sullivan K, Binns JH, Golomb BA, et al. Recent research on Gulf War illness and other health problems in veterans of the 1991 Gulf War: effects of toxicant exposures during deployment. Cortex 2016;74:449-475.


33. Lou HC. Dopamine precursors and brain function in phenylalanine hydroxylase deficiency. Acta Paediatr Suppl 1994;407:86-88.


Figure 1.
Manhattan plots and volcano plots. (A, B) Manhattan plots. Graph (A) shows p-values for CpG sites analyzed from the comparison between CMT1A patient and control groups, and (B) shows p-values for CpG sites analyzed from the comparison between de novo and non-de novo CMT1A patient groups (red dotted line, p<5×10-8; blue dotted line, p<5×10-5). (C, D) Volcano plots. Delta mean values (Δmean) were calculated based on patients or non-de novo groups (green dots, p<0.05 and Δmean≤0.2; red dots, p<0.05 and Δmean≥0.2). Graph (C) is a comparison result between patient and control groups, and (D) is a comparison result between de novo and non-de novo patient groups. CMT1A; Charcot-Marie-Tooth disease type 1A.
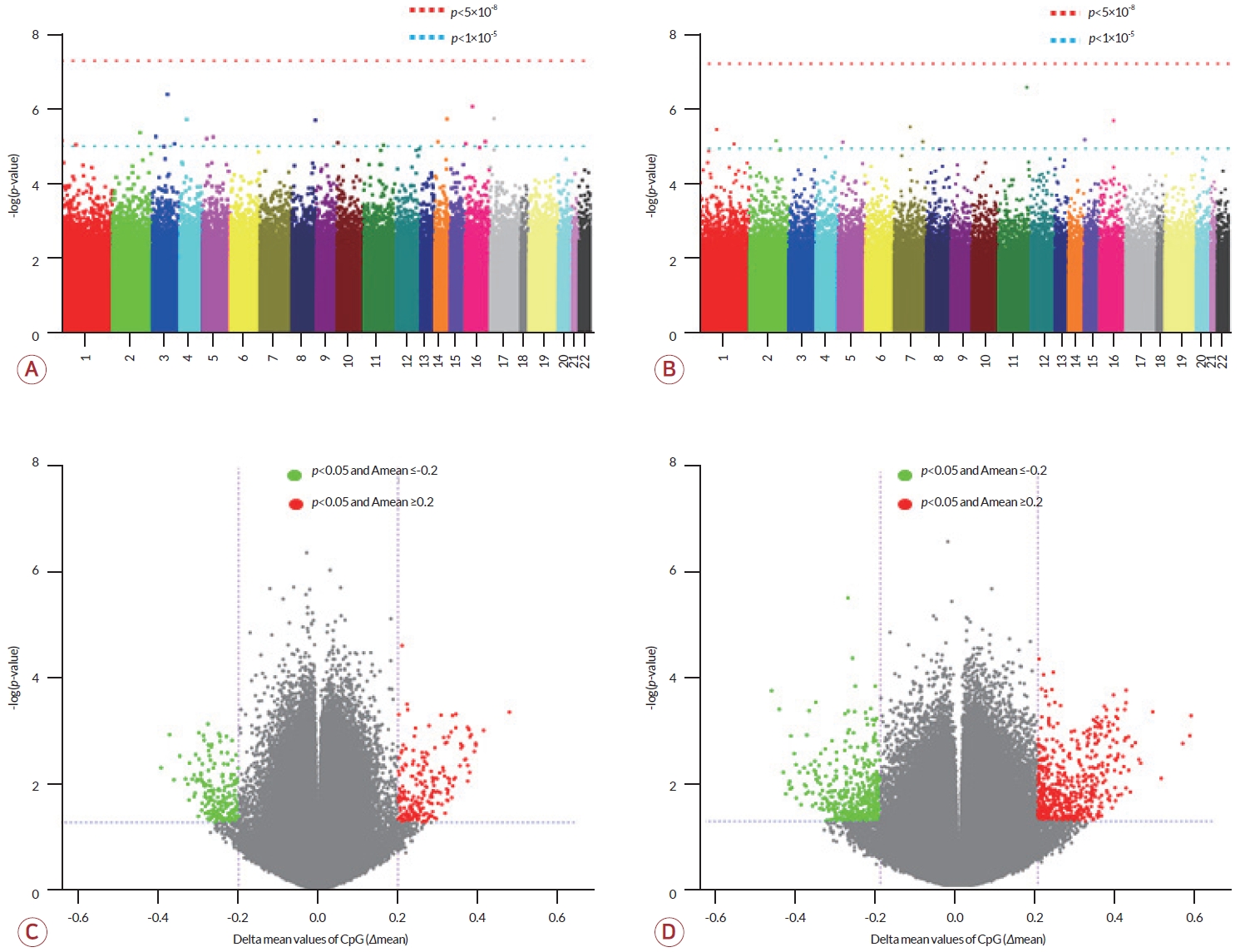
Figure 2.
Heatmaps for DMCs between comparative groups. Clustering and heatmaps were constructed according to the methylated level of each sample for DMCs with p<0.05 and ǀΔmeanǀ≤0.2. Red indicates hypermethylated CpGs and blue indicates hypomethylated CpGs. (A) CMT1A patient group vs. control group. (B) De novo CMT1A group vs. non-de novo CMT1A group. DMC; differentially methylated CpG site, CMT1A; Charcot-Marie-Tooth disease type 1A.
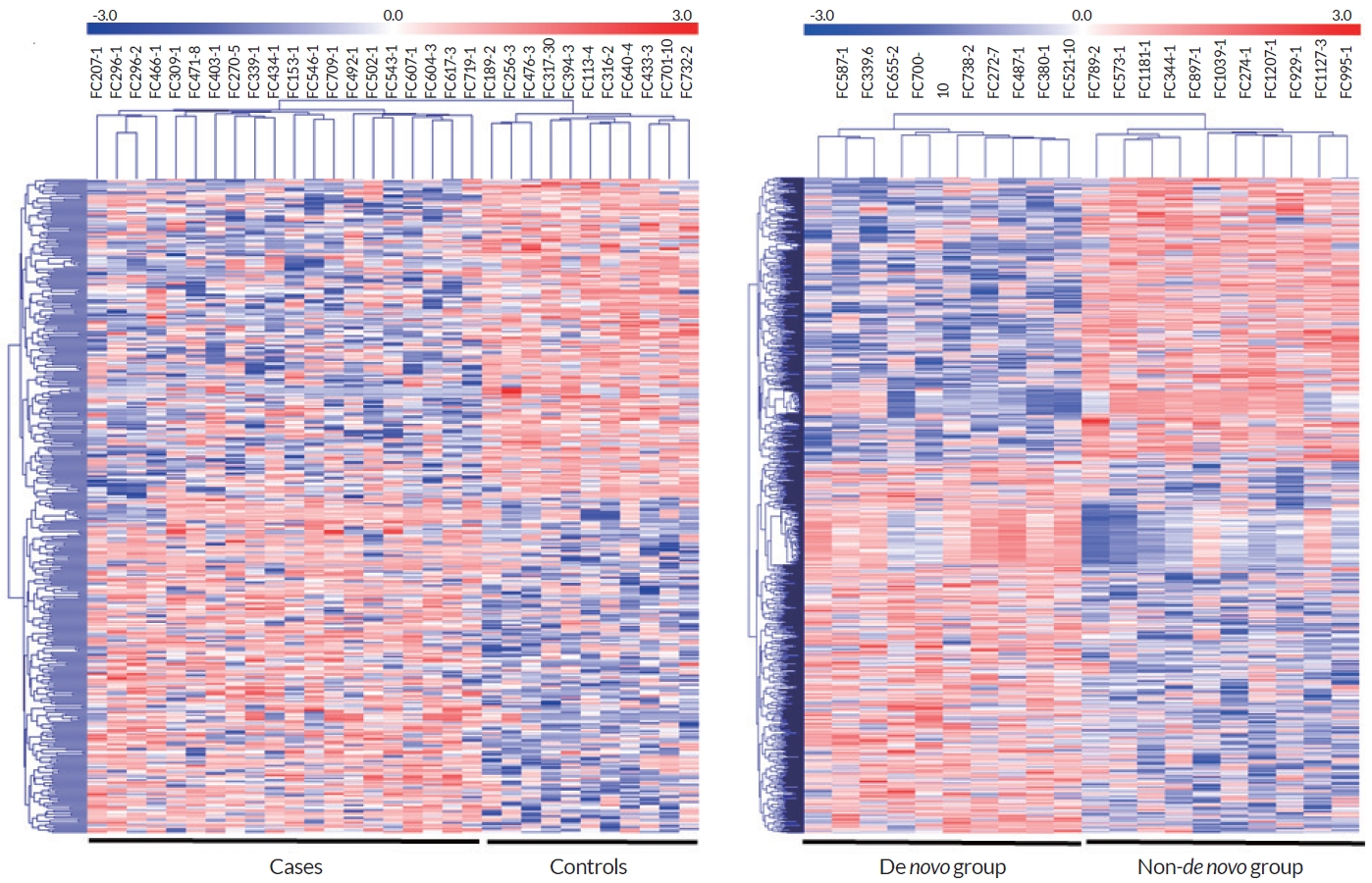
Figure 3.
Classification of major functions of genes associated with DMCs. Blue indicates GO biological process, red indicates GO molecular function, and green indicates gene classification for GO-cellular component. The values for each item were indicated as -log (p-value), and the p-value was calculated based on the ratio of DMC-related genes among all functional genes reported. (A) Patient vs. control. (B) De novo CMT1A group vs. non-de novo CMT1A group. E-box; enhancer-box, PRC1; polycomb repressive complex 1, NMDA; N-methyl-D-aspartate, DMC; differentially methylated CpG site, GO; gene ontology, CMT1A; Charcot-Marie-Tooth disease type 1A.
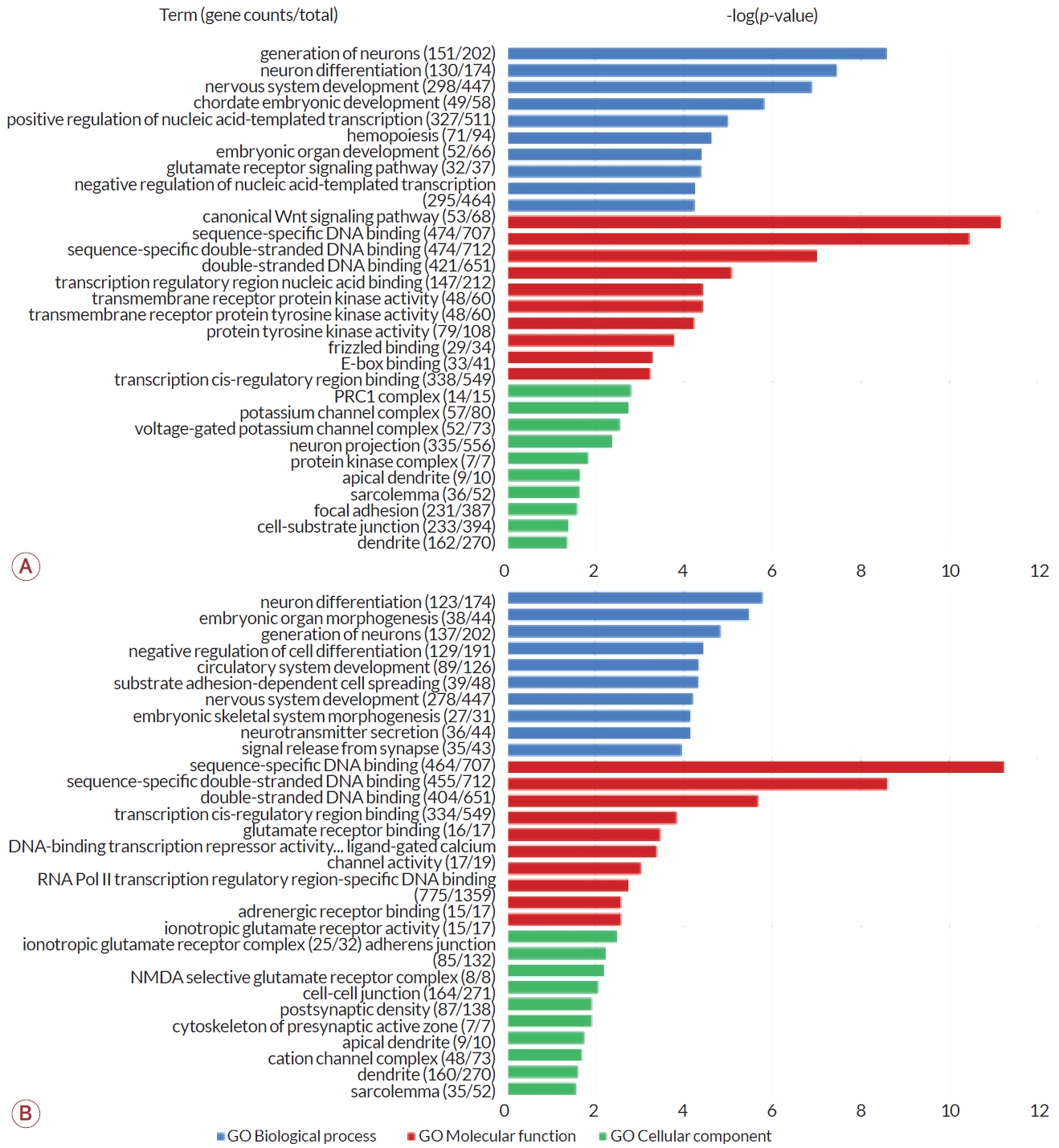
Table 1.
Comparison between patient and control groups
Table 2.
DMCs with significant differences between comparative groups (p<1×10-5)
| Group | Chromosome: position | Correlationa | Regions | Genes | p-value |
|---|---|---|---|---|---|
| Cases vs. controls | 1: 906494 | Positive | Exonic | PLEKHN1 | 7.08E-06 |
| 1: 26799761 | Negative | Intronic | HMGN2 | 9.07E-06 | |
| 2: 183732212 | Negative | Promoter | FRZB | 4.30E-06 | |
| 3: 30648879 | Negative | Intronic | TGFBR2 | 5.48E-06 | |
| 3: 123168398 | Negative | Promoter | ADCY5 | 4.02E-07 | |
| 3: 171528126 | Negative | Intronic | PLD1 | 8.59E-06 | |
| 4: 13820906 | Negative | ncRNA_intronic | LINC01182 | 1.91E-06 | |
| 5: 10331969 | Positive | Intergenic | CMBL and MARCH6 | 6.24E-06 | |
| 5: 78809625 | Negative | 5’ UTR | HOMER1 | 5.65E-06 | |
| 8: 145666083 | Negative | Intronic | TONSL | 1.98E-06 | |
| 10: 126304179 | Positive | Intergenic | LHPP and FAM53B | 8.04E-06 | |
| 11: 68671782 | Positive | Intronic | IGHMBP2b | 9.34E-06 | |
| 14: 50159769 | Negative | Promoter | KLHDC1 | 7.58E-06 | |
| 14: 102430191 | Positive | Promoter | DYNC1H1b | 1.83E-06 | |
| 16: 665214 | Negative | Intronic | RAB40C | 8.48E-06 | |
| 16: 11272371 | Positive | Exonic | CLEC16A | 8.54E-07 | |
| 16: 85688041 | positive | Exonic | GSE1 | 7.49E-06 | |
| 17: 7017905 | Negative | Promoter | ASGR2 | 1.79E-06 | |
| 22: 50277892 | Negative | Exonic | ZBED4 | 2.48E-06 | |
| 22: 50585136 | Negative | Promoter | MOV10L1 | 3.00E-06 | |
| De novo patients vs. non-de novo patients | 1: 36789537 | Negative | Promoter | EVA1B | 3.03E-06 |
| 1: 156390473 | Positive | Promoter | MIR9-1 | 7.57E-06 | |
| 2: 178483637 | Positive | 5’ UTR | TTC30A | 6.20E-06 | |
| 5: 16667385 | Negative | Intronic | MYO10 | 6.64E-06 | |
| 7: 75779880 | Negative | Intergenic | GTF2IP7 and SRRM3 | 2.60E-06 | |
| 7: 154861808 | Positive | Promoter | HTR5A | 6.57E-06 | |
| 11: 121000716 | Negative | Exonic | TECTA | 2.16E-07 | |
| 15: 25925753 | Negative | Intronic | ATP10A | 5.76E-06 | |
| 16: 54972759 | Positive | Intergenic | IRX5 and IRX6 | 1.74E-06 |
Table 3.
Stress susceptibility- or nervous system-related CpGs showing different methylation level between cases and controls in the EWAS dataseta
| Probes (Chr: position) |
Mean methylation level |
Correlation | p-value | Traits | |
|---|---|---|---|---|---|
| Cases | Controls | ||||
| cg11963436b (10: 93567261) | 0.958±0.033 | 0.928±0.041 | Positive | 0.033 | Parkinson’s disease [21] |
| cg07443748 (22: 17073594) | 0.787±0.056 | 0.830±0.041 | Negative | 0.031 | Multiple sclerosis [22] |
| cg09015566b (4: 53264) | 0.786±0.102 | 0.706±0.101 | Positive | 0.046 | Gulf War illness [19] |
| cg21006834b (13: 20138813) | 0.861±0.042 | 0.899±0.030 | Negative | 0.013 | Phelan-McDermid syndrome [23] |
| cg25040670 (6: 26022062) | 0.205±0.196 | 0.066±0.085 | Positive | 0.034 | Early social deprivation [24] |
| cg03259243b (17: 21356007) | 0.245±0.084 | 0.147±0.061 | Positive | 0.002 | Child abuse [20] |
Values are presented as mean±standard deviation.
EWAS; epigenomic wide association study, Chr; chromosome.
a EWAS Atlas (https://ngdc.cncb.ac.cn/ewas/atlas) in January 2023.
Table 4.
Stress susceptibility- or nervous system-related CpGs showing different methylation level between de novo and non-de novo groups in the EWAS dataseta
| Probes (Chr: position) |
Mean methylation level |
Correlation | p-value | Traits | |
|---|---|---|---|---|---|
| non-de novo | de novo | ||||
| cg07875360 (5:1801344) | 0.027±0.032 | 0.003±0.007 | Positive | 0.044 | Schizophrenia [25] |
| cg18303477b (10:94180046) | 0.331±0.211 | 0.160±0.120 | Positive | 0.043 | Childhood stress [11] |
| cg03144560b (20: 55043299) | 0.039±0.038 | 0.007±0.011 | Positive | 0.025 | Child abuse [20] |
| cg23144563b (6: 32157590) | 0.071±0.048 | 0.029±0.037 | Positive | 0.040 | Child abuse [20] |
| cg13904130 (6: 33085736) | 0.092±0.050 | 0.212±0.128 | Negative | 0.018 | Gulf War Illness [19] |
| cg17295225b (6: 137814971) | 0.228±0.072 | 0.146±0.093 | Positive | 0.041 | Gulf War Illness [19] |
| cg05828624b (2: 79347699) | 0.827±0.110 | 0.949±0.067 | Negative | 0.010 | Exercise [26] |
| cg02487331b (1: 146550467) | 0.774±0.210 | 0.351±0.275 | Positive | 0.001 | Ankylosing spondylitis [27] |
Values are presented as mean±standard deviation.
EWAS; epigenomic wide association study, Chr; chromosome.
a EWAS Atlas (https://ngdc.cncb.ac.cn/ewas/atlas) in January 2023.
- TOOLS
-
METRICS

-
- 0 Crossref
- 0 Scopus
- 4,810 View
- 53 Download
- Related articles
-
Molecular Genetic Analyses of Charcot-Marie-Tooth Disease Type 1A in Korean1999 ;17(6)



 PDF Links
PDF Links PubReader
PubReader ePub Link
ePub Link Full text via DOI
Full text via DOI Download Citation
Download Citation Print
Print